M. SHATSKY, P. ARBELAEZ, R. GLAESER AND S. BRENNER
JOURNAL OF STRUCTURAL BIOLOGY, 2013
Abstract
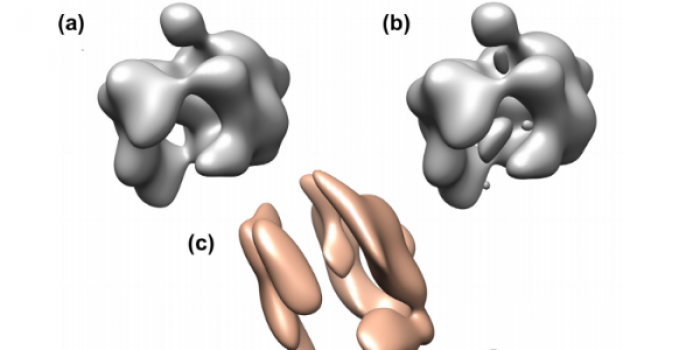
Electron tomography of intact cells has the potential to reveal the entire cellular content at a resolution corresponding to individual macromolecular complexes. Characterization of macromolecular complexes in tomograms is nevertheless an extremely challenging task due to the high level of noise, and due to the limited tilt angle that results in missing data in Fourier space. By identifying particles of the same type and averaging their 3D volumes, it is possible to obtain a structure at a more useful resolution for biological interpretation. Currently, classification and averaging of sub-tomograms is limited by the speed of computational methods that optimize alignment between two sub-tomographic volumes. The alignment optimization is hampered by the fact that the missing data in Fourier space has to be taken into account during the rotational search. A similar problem appears in single particle electron microscopy where the random conical tilt procedure may require averaging of volumes with a missing cone in Fourier space. We present a fast implementation of a method guaranteed to find an optimal rotational alignment that maximizes the constrained cross-correlation function (cCCF) computed over the actual overlap of data in Fourier space.